A prominent question in biology is how genes control the shaping or morphogenesis of an organ or an entire organism. Morphogenesis is a multifaceted process that integrates a variety of processes acting at different scales - from molecular control of gene expression to cellular coordination in a tissue. To help answer this question and better visualize morphogenesis, an interdisciplinary team of biologists, physicists, and computer scientists from the TUM and other scientific institutions built a digital image analysis pipeline based on machine learning called “PlantSeg” last year.
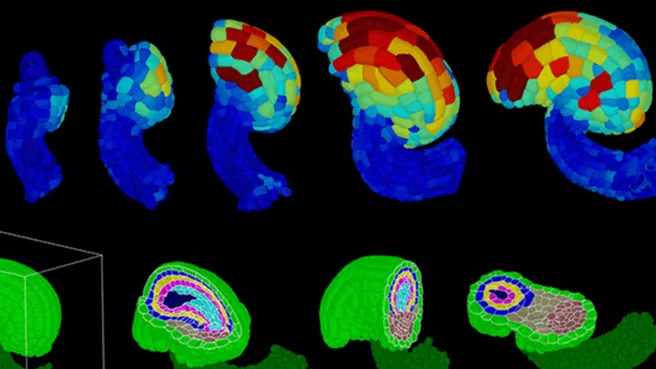
TUM doctoral students Athul Vijayan and Rachele Tofanelli from the Department of Developmental Biology of Plants (directed by Prof. Kay Schneitz) have now achieved the first successful application of PlantSeg. By using state-of the-art microscopy and the PlantSeg pipeline, they have created a digital three-dimensional atlas of ovule development in Arabidopsis thaliana. The study of its morphogenesis posed a particularly difficult challenge because the ovule has a unique curved structure combined with a complex cellular organization.
The digital atlas features complete cellular resolution even in deep tissue layers. It enables quantitative spatiotemporal analysis of cellular and gene expression patterns with cell and tissue resolution. For example, the unusual establishment of a new tissue layer in the ovule has been described for the first time, and specific cell division patterns in deeper layers have been linked to the curvature of the ovule. In addition, the cellular distribution of gene expression of an important regulatory gene has now been quantitatively described. The result of this work lays the foundation for the next generation of studies on morphogenesis.
Video:
3D digital ovules based on deep imaging, ML-based 3D segmentation, and tissue annotation.
Publication:
Vijayan, A., Tofanelli, R., Strauss, S., Cerrone, L., Wolny, A., Strohmeier, J., Kreshuk, A., Hamprecht, F. A., Smith, R. S., and Schneitz, K. (2021). A digital 3D reference atlas reveals cellular growth patterns shaping the Arabidopsis ovule. eLife 10, e63262.
Links:
Digital 3D atlas of ovule development
https://elifesciences.org/articles/63262
PlantSeg Pipeline
https://elifesciences.org/articles/57613
https://github.com/hci-unihd/plant-seg
FOR 2581
https://www.plantmorphodynamics.com
More information:
The work was carried out as part of the research group FOR 2581 "Plant Morphodynamics," which is funded by the German Research Foundation. The research group includes experts from TUM, EMBL Heidelberg, the Max Planck Institute for Plant Breeding Research in Cologne, the University of Heidelberg and the John Innes Centre in Norwich, UK. The consortium consist of biologists, physicists and computer scientists whose common interest is to understand how plants shape their organs.
Editing:
Susanne Neumann
TUM School of Life Sciences
Press- and Public relations
Scientific Contact:
Prof. Dr. Kay Schneitz
TUM School of Life Sciences
Professorship of Plant Developmental Biology
Tel. +49 8161 71 5438
kay.schneitz(at)tum.de